ITCC P4 - Paediatric Preclinical Proof Of Concept Platform
ITCC-P4: a new platform to accelerate drug development
for children and adolescents dying of cancer
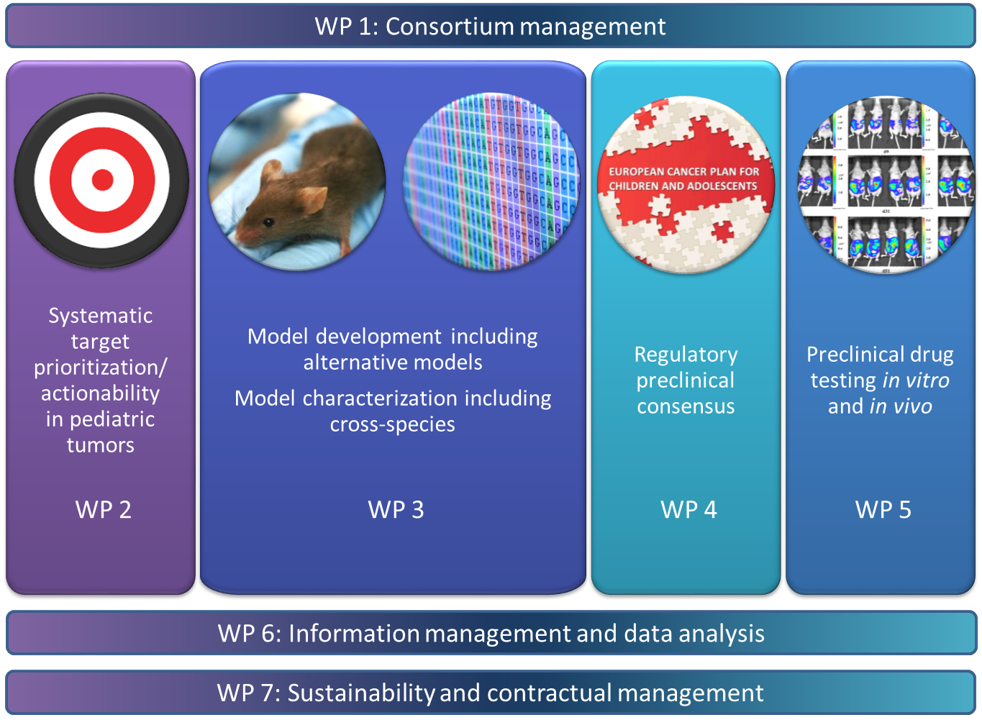
Work Packages
To facilitate the organization and management, ITCC-P4 is structured as seven work packages (WPs). Each Work Package has specific leaders (academic and pharma) who are responsible for the management and the results of their WP.
By clicking on the different WPs you can find out more about their objectives and tasks!